Haplotype networks in R
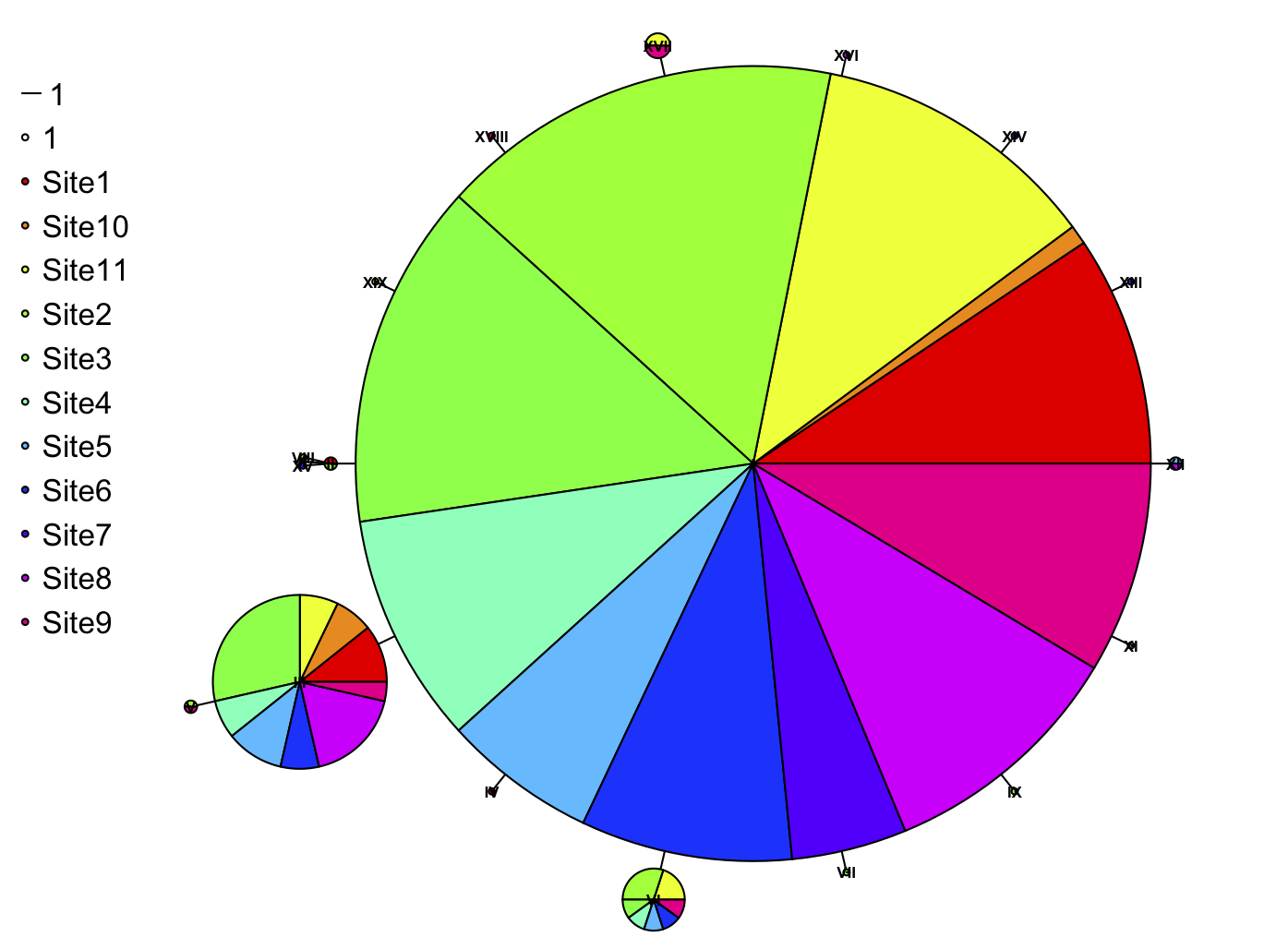
Another grad student in the lab (Kim Tenggardjaja) has been making haplotype networks like the one shown above for some of her research. While she has been calculating haplotype frequencies as well as constructing and plotting the network using the package pegas in R, there was a step in the middle (assigning individual names/sampling locations to their haplotypes) which she was doing tediously by hand in Excel. Some of my recent bumbling in R has led me to see a faster workaround using the packages plyr and reshape. I’m sure there must be a better, easier, more streamlined way of doing this, probably built right into pegas, but this solution struck me as acceptable for now. The networks may come out looking a little wonky using only pegas; I suspect the best way to gussy them up in R would be using the package igraph. The R script I wrote can be found on the resources page and here.